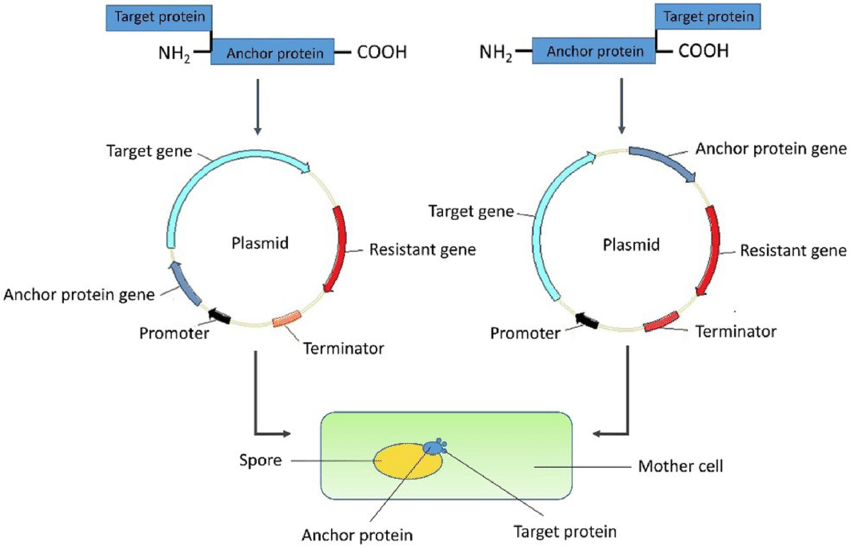
Cusabio C-terminal Myc-taggedd Recombinant
Description Myc tag monoclonal antibody CSB-MA000041M0m was produced in the immunized mouse using the synthetic peptide conjugate EQKLISEEDL (Myc) with KLH as immunogen. The Myc target peptide is a popular short peptide tag (EQKLISEEDL) derived from the c-Myc gene product…
Read more